Clinical Technoshot: in situ Tumor Cell Transcriptomes
Gioacchino Natoli
Clinical Technoshot Coordinator
| [email protected] | |
| Location |
Building 13
Floor 3rd Via Adamello 16, Milano |
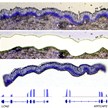
Analysis of gene expression profiles in cancer is informative of tumor properties and in some cases allows stratifying patients for diagnosis, prognostic assessment and therapeutic decisions. However, standard analyses of tumor transcriptomes average data obtained from tumor cells, which may be highly heterogeneous, and not-tumor cells, including infiltrating immune cells and stromal components. Single cell analyses provide detailed information on gene expression profiles of individual cells after dissociation of the tumor tissue, thus being not informative of the spatial relationship between tumor cells with different gene expression profiles and non-tumor components. For instance, the relationship between tumor cells with divergent gene expression profiles and vessels or other identifiable anatomical structures cannot be determined on the basis of the analysis. This technoshot aims at generating standardized procedures for digital gene expression analysis of defined tumor anatomical areas composed of a limited number of cells (10-100) obtained by laser capture microdissection. The activity of this unit is highly complementary to the Technoshot focusing on highly multiplexed in situ FISH.
References
Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I and Itzkovitz S. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. 2017. Nature. Feb 16;542(7641):352-356. Erratum in: 2017 Nature. Mar 30;543(7647):742.
Chen J, Suo S, Tam PP, Han JJ, Peng G and Jing N. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. 2017. Nat Protoc. 12(3):566-580.