Epigenetic Mechanisms in Cancer

Diego Pasini
Group Leader
| [email protected] | |
| Location |
Building 13
Floor 1st Via Adamello 16, Milano |
Scientific Focus
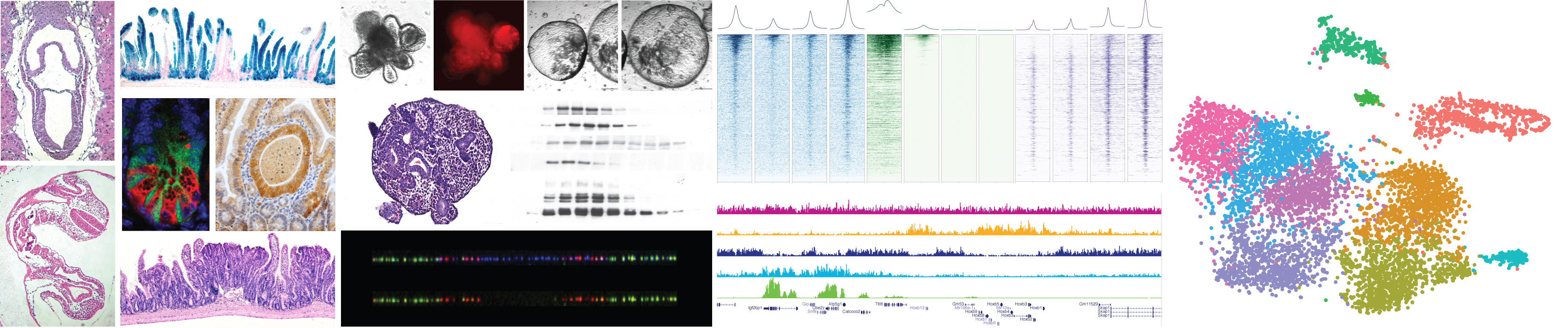
Control of cellular identity is regulated by a complex network of autonomous and non-cell autonomous signals that converge to the nucleus and instruct each individual cell to acquire specific transcription programs to exert specific functions. The coordinated activity of DNA binding transcription factors together with a plethora of chromatin modifying and remodelling enzymes is instructed to establish specific transcription programs. These mechanisms are tightly regulated and kept under control to allow proper development and or to maintain adult tissue homeostasis.
Loss of cellular identity constrains is a common feature of human tumours which frequently involves direct genetic and indirect epigenetic alterations of chromatin remodelling activities more generally defined as epigenetic factors. The central role of these activities, their enzymatic proprieties and the reduced redundancy respect to upstream signalling pathways immediately opens towards the development of novel therapeutic strategies.
The work of our laboratory is focused at characterizing the molecular mechanisms underlying distinct chromatin activities under homeostatic and pathological conditions. For this, the lab takes advantage of genetic, biochemical, transcriptomic and epigenomic approaches applied to both in vivoand 3D organoids models derived from compound mice or patient samples down to a single cell level. The work in the lab has been recently focused at:
- Defining the role of Polycomb repressive activities in adult tissue homeostasis under physiological and pathological conditions.
- Dissecting the mechanistic aspects of oncogenic mutations that targets chromatin remodelling activities.
- Characterizing the epigenetic mechanisms underlying the development of colorectal cancer.
Most Relevant Publications
-
Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, Pasini D
Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression.
Mol Cell, 2019
-
Conway E, Rossi F, Fernandez-Perez D, Ponzo E, Ferrari KJ, Zanotti M, Manganaro D, Rodighiero S, Tamburri S, Pasini D
BAP1 enhances Polycomb repression by counteracting widespread H2AK119ub1 deposition and chromatin condensation.
Mol Cell, 2021
-
Del Vecchio A, Mulé P, Fernández-Pérez D, Amato S, Lattanzi G, Zanotti M, Rustichelli S, Pivetti S, Oldani P, Mariani A, Iommazzo F, Koseki H, Facciotti F, Tamburri S, Ferrari KJ, Pasini D
PCGF6 controls murine Tuft cell differentiation via H3K9me2 modification independently of Polycomb repression.
Dev Cell, 2024
-
Tamburri S, Rustichelli S, Amato S, Pasini D
Navigating the complexity of Polycomb repression: Enzymatic cores and regulatory modules.
Mol Cell, 2024
